Difference between revisions of "Recycling Corner"
AngelHerraez (talk | contribs) (moving to proper subsection) |
AngelHerraez (talk | contribs) (moving long sections to subpages) |
||
| Line 76: | Line 76: | ||
The code below was used to create the palettes shown above. It's a pure javascript color picker that can be used for changing color of selected part of molecule, applet background, or any feature of the model (cartoons, surfaces, etc.). | The code below was used to create the palettes shown above. It's a pure javascript color picker that can be used for changing color of selected part of molecule, applet background, or any feature of the model (cartoons, surfaces, etc.). | ||
| − | Sample of use | + | Sample of use, JavaScript code needed, example page: [[Recycling_Corner/Molecule_color_picker]] |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | JavaScript code needed | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=== "Basic" spin toggle === | === "Basic" spin toggle === | ||
Revision as of 13:57, 23 October 2013
This section intends to collect material by some users that may be useful to other users.
Feel free to add your material!
- Websites Using J(S)mol
- J(S)mol in the Classroom
- Journals Using J(S)mol
- J(S)mol Literature
- Articles Using J(S)mol
- Books Using J(S)mol
- Wikis Using J(S)mol
- Blogs Using J(S)mol
- CMS Using J(S)mol (Moodle and others)
Course Management Systems, Learning Management Systems, Virtual Learning Environments, e-Learning Platforms - Applications Embedding Jmol
- Distributions Including Jmol
- Blue Obelisk
For example:
Contents
- 1 Jmol logos, banners, icons
- 2 Templates for creating Jmol webpages
- 3 Reusable scripts
- 3.1 Molecule color picker
- 3.2 "Basic" spin toggle
- 3.3 "Foolproof" spin toggle
- 3.4 Slider controls for Jmol
- 3.5 Echoing PDB "Title" to the applet window
- 3.6 Providing a 'please wait' notice while the applet loads
- 3.7 Exporting an image from the applet
- 3.8 Opening a duplicate of the model in a resizable pop-up window
- 3.9 Style transitions
- 3.10 Replacing applet with signed applet upon demand
- 3.11 Including special characters in echo
- 3.12 Center of mass
- 3.13 RIBOZOME - an Alpha Helix Generator script
- 4 Custom pop-up menus
Jmol logos, banners, icons
New 2013 Jmol icon and logos
Icon, png:
Windows icon; .ico files cannot be shown on this webpage, but you can download them.
for more icon formats, visit Angel's webpage
Logos and banners:
'Made with Jmol' banners, in several languages: Jmol_Images
Older versions
Templates for creating Jmol webpages
Jmol_Web_Page_Maker
Jonathan Gutow has written a Java program that automates generation of webpages that include Jmol. The two templates below are among the types of web pages that can be generated. When using Jmol_Web_Page_Maker for the templates below, you can choose the view(s) to be displayed inside the Jmol application and have the necessary scripts, images plus .html code generated and saved automatically. Visit his website.
This is now superceeded by the "export to web page" ability included within recent versions of Jmol application (11.3.x).
Easy assignments for students or quick building of a simple page!
SChiSM2
SChiSM2 is a program running in a web server that lets you easily create a web page that includes interactive molecular models using JmolApplet.
- SChiSM2 home page
- A description has been published:
- S. Cammer (2007) SChiSM2: creating interactive web page annotations of molecular structure models using Jmol. Bioinformatics 23:383-384. doi:10.1093/bioinformatics/btl603 [1]
Jmol "pop-up" and "pop-in"
Delays the appearance of Jmol panel until the user requests it. An image or caption is shown and upon user's choice it is replaced by the applet.
No more waiting for Java and Jmol to load if you are not interested in the 3D model!
Two methods have been developed:
- Method A - versatile: The user clicks to see the 3D model, and chooses whether it will be opened inline within the page (âpop-inâ), or in a common pop-up window, or in separate pop-up windows for each model. (Uses <iframe>.)
- Method B - simpler: The user clicks to see the 3D model, always inline within the page (âpop-inâ). (Uses <div>.) (A similar method is used on Jmol home webpage.)
You can find more details, demo pages and downloadable templates for both methods at Biomodel website.
For an alternative method, see Bill Reusch's page that uses innerHTML.
For other pop-up solutions, see also below.
Dynamically resized panels layout
Six templates for dividing the screen into two or more panels, either for Jmol or for content, all sized in percent relative to window size, using the whole window space, and automatically resized when the window is resized; no scrollbars in Jmol section, scrollbars in content if needed, no duplicate parallel scrollbars.
No more guessing the user's screen resolution, browser chrome, default font size!
You can find more details, demo pages and downloadable templates at Biomodel website.
Reusable scripts
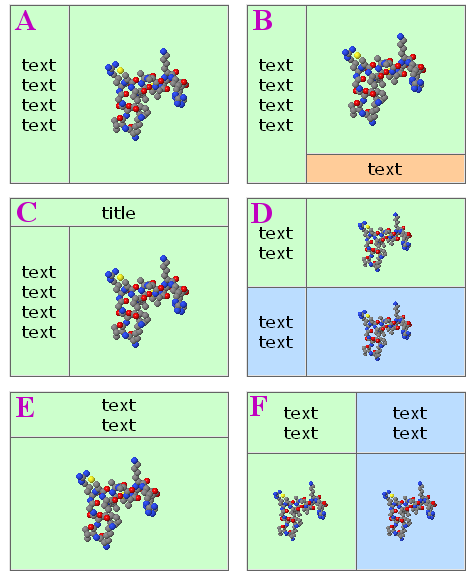
Molecule color picker
The code below was used to create the palettes shown above. It's a pure javascript color picker that can be used for changing color of selected part of molecule, applet background, or any feature of the model (cartoons, surfaces, etc.).
Sample of use, JavaScript code needed, example page: Recycling_Corner/Molecule_color_picker
"Basic" spin toggle
Quite trivial, but compact and useful. However, if other scripts, controls, or the user change the spin status, the checkbox will need to be reset accordingly using javascript. (To avoid this limitation, see the "foolproof" solution below)
It will work with any version of Jmol (as long as Jmol.js is being used in the web page).
jmolCheckbox("spin on", "spin off", "spin", false)
if the model is initially not spinning (the default);
or
jmolCheckbox("spin on", "spin off", "spin", true)
if the model is initially spinning (due to another script).
(The word "spin" above can be changed to any other text you like.)
"Foolproof" spin toggle
Reads the current spin status from the applet and changes it, so it can cope with any other previous change via script, popup menu, page controls, etc.
The best and simplest trick:
Valid for recent versions of Jmol:
jmolButton("if(_spinning);spin off;else;spin on;endif","toggle spin")
(If you prefer jmolLink, it can be used in the same manner. If you don't use Jmol.js, and you have a good reason for not using it, you will know how to modify this code.)
(The words "toggle spin" above can be changed to any other text you like.)
The old trick:
Only compatible with Jmol 11.x (as long as Jmol.js is being used in the web page).
jmolButton("show spin", "toggle spin")
(If you prefer jmolLink, it can be used in the same manner.)
(The words "toggle spin" above can be changed to any other text you like.)
You must include these additional pieces of code:
Somewhere between <script> tags (recommended: within the head section):
function messageProcess(a_,b_)
{ // Convert both parameters from Java strings to JavaScript strings:
var a = (""+a_).substring("jmolApplet".length)
var b = ""+b_
// a_ will start with "jmolApplet": strip and leave just the ID part
/* SPIN DETECTION AND TOGGLE */
if ( b.indexOf("set spin") != -1 )
{ if ( b.indexOf("spin on") != -1 ) //was spinning
{ jmolScript("spin off", a)
}
else //was not spinning
{ jmolScript("spin on", a)
}
}
/* END of SPIN DETECTION AND TOGGLE */
}
and somewhere between <script> tags, after the call to jmolInitialize() and before the call to jmolApplet():
jmolSetCallback("messageCallback", "messageProcess")
(The word "messageProcess" above can be changed to any other text you like, as long as it is the same in the above two sections of code.)
Slider controls for Jmol
Version (A)
This has an amazing look-and-feel. Uses javascript, Jmol.js and Walter Zorn's Drag'n'Drop & DHTML Library, © Walter Zorn under GNU LGPL.
See it in action.
Steps to follow:
- Download Walter Zorn's library from http://www.walterzorn.com/dragdrop/dragdrop_e.htm (You need the
wz_dragdrop.jsfile.) - Get a copy of the image files,
track.gifand
sliderthumb.gif. You could use your own, but size is important since the code is related to it. - Include this code in your webpage:
<head>
<script src="../../Jmol.js" type="text/javascript"></script>
<script type="text/javascript">
/* Uses Walter Zorn's Drag'n'Drop & DHTML Library (GNU LGPL).
Integration with Jmol by Angel Herráez.
*/
function slabSlider()
{
jmolScript( 'slab ' + eval(100-(20+dd.elements.thumb.x-dd.elements.thumb.defx)) )
document.getElementById("posi").innerHTML = 100-(20+dd.elements.thumb.x-dd.elements.thumb.defx)
}
</script>
</head>
<body>
<script type="text/javascript" src="wz_dragdrop.js"></script>
<script type="text/javascript">
jmolInitialize("../../")
jmolApplet(300, "load lysozyme.pdb; spacefill;")
</script>
<input type="button" value="prepare for slabbing"
onClick='jmolScript("spin off; slab on; slab 80;"); document.getElementById("posi").innerHTML="80"; dd.elements.thumb.moveTo(dd.elements.track.x+20, dd.elements.track.y); '>
<br>
<img name="track" src="track.gif" width="104" height="29" alt="" title="" align="absmiddle">
<img name="thumb" src="sliderthumb.gif" width="18" height="29" alt="" title="move me" align="absmiddle">
<span id="posi">80</span>%
<br>
<input type="button" value="cancel slabbing"
onClick='jmolScript("slab off;")'>
<script type="text/javascript">
/* Courtesy of Walter Zorn;
Drag'nDrop & DHTML Library (c)Walter Zorn under the LGPL (Lesser General Public License, http://www.gnu.org/copyleft/lesser.html
*/
SET_DHTML("thumb"+MAXOFFLEFT+20+MAXOFFRIGHT+80+HORIZONTAL,"track"+NO_DRAG)
dd.elements.thumb.moveTo(dd.elements.track.x+20, dd.elements.track.y);
dd.elements.thumb.setZ(dd.elements.track.z+1);
dd.elements.track.addChild("thumb");
dd.elements.thumb.defx = dd.elements.track.x+20;
dd.elements.thumb.setDragFunc(slabSlider);
//dd.elements.thumb.setDropFunc(slabSlider); //alternative, if it cannot cope with the other
</script>
</body>
Important Notes:
- You must adjust the path to
Jmol.js,jmolInitialize()andwz_dragdrop.jsaccording to your set-up. - The order of the code element in the above example is crucial. In particular, wz_dragdrop.js must be included in the body, not in the head. If in the head, the slider works in Firefox and Safari, but not in Internet Explorer. Also, the block starting with SET_DHTML(...) must follow the block that displays the slider itself on the page.
Version (B)
This looks excellent too, and has some more versatility. Uses javascript, CSS, Jmol.js and WebFX Slider, © Erik Arvidsson under Apache Software License.
See instructions and several examples here, applied to slabbing, bond thickness, translucency, ribbon width and thickness.
Echoing PDB "Title" to the applet window
The following JavaScript code snippet will report the title of a PDB entry to the applet window when a PDB file is loaded. This should be included within a JavaScript portion of the html file that loads the applet and the PDB file. One caveat: the "TITLE" in a PDB file is depositor-generated and may or may not be "useful" to the end user! :)
var headerInfo = jmolGetPropertyAsString("fileHeader");
var cutUp = headerInfo.split("\n");
var headerstring="";
for (l=0;l<cutUp.length;l++) {
var regexp = new RegExp("TITLE.{5}(.*)\s*");
var temp = cutUp[l];
if (temp.search(regexp) == 0) {
temp2 = RegExp.$1;
temp2.replace(/^\s+|\s+$/g, "").replace(/\s+/g, " ");
temp2 = temp2 + "|";
headerstring += temp2; }
}
}
headerstring.replace(/TITLE/, "");
jmolScript("set echo depth 0; set echo headerecho 2% 2%; font echo 12 sanserif bolditalic; color echo green");
jmolScript('echo "' + headerstring + '"');
Bob Hanson has also contributed this much more elegant way to accomplish the same thing all within JmolScript:
function getHeader()
var titleLines = getProperty("fileHeader").split().find("TITLE").split()
for (var i = 1; i <= titleLines.size;i = i + 1)
titleLines[i] = (titleLines[i])[11][0].trim()
end for
return titleLines.join("|")
end function
set echo bottom left
echo @{getheader()}
Providing a 'please wait' notice while the applet loads
in a separate page
Exporting an image from the applet
An image of the current view may be exported from the Jmol applet, either
- in a pop-up window, also compatible with JSO and JSmol.
- inserted into the same web page
Opening a duplicate of the model in a resizable pop-up window
The model, with is current state, is duplicated or cloned into a new (pop-up) window so it may be displayed at a larger size (and the user may resize that window at will).
The classic version (not shown here) is good for Jmol.js-based JmolApplet.
This version is for JSmol (in either HTML5, Java or WebGL modalities).
You need a portion of JavaScript code included in your page and a separate html file (for the pop-up), also with the relevant code.
1. Code in your page:
<script type="text/javascript">
var JSmolCloneData = {};
function cloneJSmol(JSmolObject) {
var t = JSmolObject._jmolType; //temp
if ( /_Canvas2D/.test(t) ) { t = 'HTML5'; }
else if ( /_Canvas3D/.test(t) ) { t = 'WebGL'; }
else if ( /_Applet/.test(t) ) { t = 'Java'; }
else { t = null; }
JSmolCloneData.type = t;
JSmolCloneData.state = Jmol.getPropertyAsString(JSmolObject, 'stateInfo');
window.open('JSmolPopup.htm','JSmolPopup','resizable, width=500, height=500');
}
</script>
<input type="button" value="clone JSmol in a popup window" onClick="cloneJSmol(myJmol)">
Note: myJmol should be changed to whatever you have named your JSmol object - and it is the object itself, not a text string.
2. Source code of the ![]() JSmolPopup.htm file:
JSmolPopup.htm file:
<!DOCTYPE html>
<html>
<title>cloned JSmol</title>
<head>
<meta charset="utf-8">
<style type="text/css">
html , body { height:100%; overflow:hidden; margin:0; padding:0; }
</style>
<script type="text/javascript" src="JSmol.min.js"></script>
<script type="text/javascript">
if (opener.JSmolCloneData.type == 'WebGL') {
document.writeln('<script src="js/JSmolThree.js" type="text/javascript"><'+'/script>');
document.writeln('<script src="js/JSmolGLmol.js" type="text/javascript"></'+'script>');
}
</script>
</head>
<body>
<script type="text/javascript">
var cloneInfo = {
height: '100%',
width: '100%',
isSigned: false,
jarFile: 'JmolApplet0.jar',
jarPath: 'java',
j2sPath: 'j2s',
script: opener.JSmolCloneData.state.replace(/zoomLarge true/i,'zoomLarge false'),
use: opener.JSmolCloneData.type
};
Jmol.getApplet("JSmolClone", cloneInfo);
</script>
</body>
Note: you need to adjust the paths to ![]() JSmol.min.js,
JSmol.min.js, ![]() j2s and
j2s and ![]() java
java
Style transitions
Smooth transitions from one rendering style to another.
Spacefill to ball & stick
for (var i=0;i<=9;i++) {
s=100-80*i/9;
spacefill @s%;
w=0.9-0.75*i/9;
wireframe @w;
delay 0.2;
}
Can be given in a single line:
for(var i=0;i<=9;i++){s=100-80*i/9;spacefill @s%;w=0.9-0.75*i/9;wireframe @w;delay 0.2;}
Replacing applet with signed applet upon demand
The signed applet has certain capabilities that the unsigned applet lacks, but using the signed applet will raise some dialogs with security warnings that the general user may find scary and, if permission is denied, the expanded features will not be available and hence the page will fail to do what is expected.
This may lead to a situation where webpage authors wish to use the unsigned applet by default and make the switch to the signed applet if and when the users requests a certain capability (like saving a model to disk). The code here will do that.
The rationale is:
- Store the current state of the model in the applet so that it can be restored after the applet switch.
- Destroy the applet object.
- Insert the signed applet in the same place.
- Load back the model and state.
Latest Jmol.js (post-26 Sep 2011, Jmol_12.2.RC8 or later) includes a new jmolSwitchToSignedApplet() function to do this replacement easier:
The code (html + JavaScript, relying on Jmol.js):
<script type="text/javascript">
jmolInitialize(".");
jmolApplet(400, 'load example.mol;', 'ABC');
</script>
<br>
<input type="button" value="make the switch" onClick="jmolSwitchToSignedApplet('ABC')">
That's all! Note that 'ABC' is an example of an applet suffix ID, and if omitted will default to '0' (zero) in both jmolApplet() and jmolSwitchToSignedApplet().
Warning!
- This method has not been thoroughly tested and might fail or break the page; particularly, testing under different browsers and OS's is needed.
Including special characters in echo
Non-keyboard characters may be included in echo texts using their Unicode character number, like this:
echo "\u03B1 is the alpha letter"; echo "\u2192 is an arrow pointing right"; echo "\u00B2 is the superscript two, or square power";
Center of mass
Jmol centers the model around its geometric center, i.e. the center of the boundbox. If you are interested in the mass center of the molecule:
1. Run or define or load from a script file this content in JmolScript:
function CoM(atomSet) {
if (atomSet=="") {
print "Warning: there are no atoms in the provided set. Using all atoms in the model.";
atomSet = {*};
}
var n = atomSet.size;
var mx = 0; var my = 0; var mz = 0;
for (i=0; i<n; i+=1) {
mx = mx + atomSet[i].x * atomSet[i].mass;
my = my + atomSet[i].y * atomSet[i].mass;
mz = mz + atomSet[i].z * atomSet[i].mass;
}
var m = atomSet.mass.sum;
mx = mx / m;
my = my / m;
mz = mz / m;
return {@mx @my @mz};
}
function drawCoM(atomSet) {
draw ctr diameter 0.9 color translucent yellowTint @{CoM(atomSet)};
}
function axesCoM(atomSet) {
axes center @{CoM(atomSet)};
axes on;
}
2. after that has been executed, you can
- retrieve the coordinates of the center:
c = CoM(); // using all atoms
print @c;
c = CoM( {_C},{_O} ); // just the carbon and oxygen atoms
print @c;
- draw a sphere in the center of mass:
drawCoM();
drawCoM( {_C},{_O} );
- place axes in the center of mass:
axesCoM();
axesCoM( {_C},{_O} );
RIBOZOME - an Alpha Helix Generator script
Creates an alpha helix from a user input string.
# RIBOZOME - Jmol script by Ron Mignery with help from Dr. Angel Herráez
# v1.2 beta 10/23/2013
#
# New in v1.2 beta:
# No longer zaps existing atoms
# Now adds on to existing helix on subsequent runs
# Fixes proline cross-link bug with XXXXPXP
# Localizes variables and globalizes constants
#
# RIBOZOME takes a string message encoding an amino acid (aa) sequence
# and generates a corresponding alpha helix one aa at a time from the
# N terminus to the C terminus rotating the emerging helix as it goes.
#
# The message is a string entered by the user at a prompt.
# It may be typed in or pasted in and be of any length
# If the message is prepended with <C>: (where C is any single letter)
# then the chain is so labeled and separated from existing chains
# if different from the first chain.
#
# The IUPAC/IUBMB 1 letter code is used:
# A=ALAnine B=GLutam?X* C=CYSteine D=ASPartate E=GLUtamate
# F=PHEnylalanine G=GLYcine H=HIStidine I=IsoLEucine K=LYSine
# L=LEUcine M=METhionine N=ASparagiNe O=PYrroLysine*** P=PROline
# Q=GLutamiNe R=ARGinine S=SERine T=THReonine U=SElenoCysteine
# V=VALine W=TRyPtophan X=UNKnown Y=TYRosine Z=ASpar?X**
# *Either GLU or GLN: drawn as GLN with chi3 flipped
# **Either ASP or ASN: drawn as ASN with chi3 flipped
# ***Not supported: drawn as ALA
# The following constant values determine the pitch of the alpha helix
gPHI = -57 # Dihedral angle of N-CA bond (nominally -57)
gPSI = -47 # Dihedral angle of CA-C bond (nominally -47)
gOMEGA = 180 # Dihedral angle of the peptide bond (nominally 180)
gPEPTIDE_ANGLE = 110 # C-N-CA angle (nominally 110)
gPRO_BUMP = -10 # Psi angle change increment to N-3psi when N is Pro
gCHAIN = 'A' # The chain id
# Lookup 3 letter code from 1 letter code
g3from1 = {"A":"ALA", "B":"GLX","C":"CYS", "D":"ASP","E":"GLU", "F":"PHE",
"G":"GLY", "H":"HIS","I":"ILE", "K":"LYS","L":"LEU", "M":"MET",
"N":"GLN", "O":"PYL","P":"PRO", "Q":"GLN","R":"ARG", "S":"SER",
"T":"THR", "U":"SEC","V":"VAL", "W":"TRP","X":"UNK", "Y":"TYR", "Z":"ASX"}
# Generate PDB atom record
function genAtom(e, aa, i, xyz) {
gA = format("ATOM %5d %4s %3s ", gN, e, aa )
gA += format("%s%4d %8.3f", gCHAIN, i, xyz[1] )
gA += format("%8.3f%8.3f\n", xyz[2], xyz[3] )
gn++
return gA
};
# Generate a PDB amino acid record set
function genAA(i, aa) { # Writes globals gA and gN
# From constructed AAs
var N0 = [0.0, 0.0, 0.0]
var CA = [ 0.200, 1.174, 0.911 ]
var C = [ -1.110, 1.668, 1.425 ]
var O = [ -1.320, 1.693, 2.62 ]
var CB = [ 1.062, 2.1950, 0.230 ]
var G1 = [ 2.359, 1.607, -0.344]
var G2 = [ 1.363, 3.336, 1.157 ]
var D1 = [ 3.222, 2.656, -1.048 ]
var D2 = [ 3.143, 0.904, 0.725 ]
var E1 = [ 3.645, 3.749, -0.167 ]
var E2 = [ 2.491, 3.234, -2.249 ]
var Z = [ 4.470, 4.717, -0.885 ]
var H1 = [ 4.450, 6.006, -0.220 ]
var H2 = [5.833, 4.228, -0.984 ]
var Gp = [ 2.008, 1.24, -0.46 ]
var Dp = [1.022, 0.213, -1.031 ]
var Gfy = [ 2.368, 1.471, -0.0152 ]
var D1fy = [ 3.346, 1.524, 0.921 ]
var D2fy = [ 2.493, 0.516, -1.151 ]
var E1fy = [ 4.513, 0.615, 0.8244 ]
var E2fy = [ 3.528, -0.336, -1.206 ]
var Zfy = [ 4.588, -0.285, -0.168 ]
var Hfy = [ 5.738, -1.245, -0.233 ]
var Ghw = [ 2.406, 1.626, -0.134 ]
var D1hw = [3.498, 1.936, 0.675]
var D2hw = [ 2.713, 0.901, -1.211 ]
var E1hw = [ 4.160, 0.518, -1.178 ]
var E2hw = [ 4.622, 1.160, 0.0816 ]
var E3hw = [ 3.789, 2.523, 1.944 ]
var Z2hw = [ 5.973, 1.177, 0.689 ]
var Z3hw = [ 5.014, 2.550, 2.503 ]
var H2hw = [ 6.153, 1.846, 1.844 ]
#N1 = [ 2.069, -2.122, -0.554]
# Build PDB atom records common to all AAs
a3 = g3from1[aa]
if (a3 = "") {
a3 = "UNK"
}
print format("+ %s%d/%d", a3, i, gSeq.count + gResno)
gA = genAtom(" N ", a3, i, N0)
gA += genAtom(" CA ", a3, i, CA)
gA += genAtom(" C ", a3, i, C)
gA += genAtom(" O ", a3, i, O)
if ((aa != 'G') && (aa != 'X')) {
gA += genAtom(" CB ", a3, i, CB)
}
# Now add AA specific atom records
switch (aa) {
case 'A' :
break;
case 'B' :
gA += genAtom(" CG ", a3, i, G1)
gA += genAtom(" CD ", a3, i, D1)
gA += genAtom(" OE1", a3, i, E2) # GLN with Es switched
gA += genAtom(" NE2", a3, i, E1)
break;
case 'C' :
gA += genAtom(" SG ", a3, i, G2)
break;
case 'D' :
gA += genAtom(" CG ", a3, i, G1)
gA += genAtom(" OD1", a3, i, D1)
gA += genAtom(" OD2", a3, i, D2)
break;
case 'E' :
gA += genAtom(" CG ", a3, i, G1)
gA += genAtom(" CD ", a3, i, D1)
gA += genAtom(" OE1", a3, i, E1)
gA += genAtom(" OE2", a3, i, E2)
break;
case 'F' :
gA += genAtom(" CG ", a3, i, Gfy)
gA += genAtom(" CD1", a3, i, D1fy)
gA += genAtom(" CD2", a3, i, D2fy)
gA += genAtom(" CE1", a3, i, E1fy)
gA += genAtom(" CE2", a3, i, E2fy)
gA += genAtom(" CZ ", a3, i, Zfy)
break;
case 'G' :
break;
case 'H' :
gA += genAtom(" CG ", a3, i, Ghw)
gA += genAtom(" ND1", a3, i, D1hw)
gA += genAtom(" CD2", a3, i, D2hw)
gA += genAtom(" CE1", a3, i, E2hw)
gA += genAtom(" NE2", a3, i, E1hw)
break;
case 'I' :
gA += genAtom(" CG1", a3, i, G1)
gA += genAtom(" CG2", a3, i, G2)
gA += genAtom(" CD1", a3, i, D1)
break;
case 'K' :
gA += genAtom(" CG ", a3, i, G1)
gA += genAtom(" CD ", a3, i, D1)
gA += genAtom(" CE ", a3, i, E1)
gA += genAtom(" NZ ", a3, i, Z)
break;
case 'L' :
gA += genAtom(" CG1", a3, i, G1)
gA += genAtom(" CD1", a3, i, D1)
gA += genAtom(" CD2", a3, i, D2)
break;
case 'M' :
gA += genAtom(" CG ", a3, i, G1)
gA += genAtom(" SD ", a3, i, D1)
gA += genAtom(" CE ", a3, i, E1)
break;
case 'N' :
gA += genAtom(" CG ", a3, i, G1)
gA += genAtom(" OD1", a3, i, D1)
gA += genAtom(" ND2", a3, i, D2)
break;
case 'P' :
gA += genAtom(" CG ", a3, i, GP)
gA += genAtom(" CD ", a3, i, DP)
break;
case 'Q' :
gA += genAtom(" CG ", a3, i, G1)
gA += genAtom(" CD ", a3, i, D1)
gA += genAtom(" OE1", a3, i, E1)
gA += genAtom(" NE2", a3, i, E2)
break;
case 'R' :
gA += genAtom(" CG ", a3, i, G1)
gA += genAtom(" CD ", a3, i, D1)
gA += genAtom(" NE ", a3, i, E1)
gA += genAtom(" CZ ", a3, i, Z)
gA += genAtom(" NH1", a3, i, H1)
gA += genAtom(" NH2", a3, i, H2)
break;
case 'S' :
gA += genAtom(" OG ", a3, i, G1)
break;
case 'T' :
gA += genAtom(" OG1", a3, i, G1)
gA += genAtom(" CG2", a3, i, G2)
break;
case 'U' :
gA += genAtom("SeG ", a3, i, G1)
break;
case 'V' :
gA += genAtom(" CG1", a3, i, G1)
gA += genAtom(" CG2", a3, i, G2)
break;
case 'W' :
gA += genAtom(" CG ", a3, i, Ghw)
gA += genAtom(" CD1", a3, i, D1hw)
gA += genAtom(" CD2", a3, i, D2hw)
gA += genAtom(" CE2", a3, i, E2hw)
gA += genAtom(" NE1", a3, i, E1hw)
gA += genAtom(" CE3", a3, i, E3hw)
gA += genAtom(" CZ2", a3, i, Z2hw)
gA += genAtom(" CZ3", a3, i, Z3hw)
gA += genAtom(" CH2", a3, i, H2hw)
break;
case 'X' :
gA += genAtom(" Xx ", a3, i, CB)
break;
case 'Y' :
gA += genAtom(" CG ", a3, i, Gfy)
gA += genAtom(" CD1", a3, i, D1fy)
gA += genAtom(" CD2", a3, i, D2fy)
gA += genAtom(" CE1", a3, i, E1fy)
gA += genAtom(" CE2", a3, i, E2fy)
gA += genAtom(" CZ ", a3, i, Zfy)
gA += genAtom(" OH ", a3, i, Hfy)
break;
case 'Z' :
gA += genAtom(" CG ", a3, i, G1)
gA += genAtom(" OD1", a3, i, D2) # ASN with Ds switched
gA += genAtom(" ND2", a3, i, D1)
break;
default :
break;
}
return gA
};
# Generate an alpha helix
function genAlpha(gSeq) {
gN = all.count + 1 # global new N atom index
# Find last linkable N if any
gResno = 0 # global pre-existing AA count
var pn = 1 # previous gN
for (var i = all.count-1; i > 0; i--) {
# If found
if (distance({atomno=i}, {0,0,0}) < 0.1) {
pn = i
# If new chain, separate from existing chain
if ({atomno=i}.chain != gCHAIN) {
select all
translateselected {2.069, -2.122, -0.554 } #N1
}
else {
gResno = {atomno=i}.resno
}
break;
}
}
# For each aa
set appendnew false
var nn = gN # new N
for (var i = 1; i <= gSeq.count; i++) {
# Move polypeptide C to bond distance from new AA N
select all
fix none
translateselected {2.069, -2.122, -0.554 } #N1
# Gen AA
gA = "data \"append aa\"\n" # global PDB atom record
gA += genAA(i + gResno, gSeq[i]); # gN is updated in subroutine
gA += "end \"append aa\""
script inline @{gA}
# If PRO ahead
var pb = 0
if ((gSeq.count - i) >= 2) {
if (gSeq[i + 2] == 'P') {
pb = gPRO_BUMP
}
}
# If not first AA
if (nn > 1) {
# Gen axis on new N perpendicular to the plane
# containing atoms nn, pn+2 and nn+1
var v1={atomno = @{pn+2}}.xyz - {atomno = nn}.xyz
var v2={atomno = @{nn+1}}.xyz - {atomno = nn}.xyz
var axis = cross(v1, v2)
# Center on atom previous C
axis += {atomno = @{pn+2}}.xyz
# Rotate the polypeptide N on the new AA C to tetrahedral (nominally 110)
select atomno < nn
fix atomno >= nn
rotateselected @axis {atomno = nn} @{gPEPTIDE_ANGLE - 69.3}
# Make omega dihedral = gOMEGA (nominally 180)
rotateselected {atomno=@{pn+2}} {atomno=nn} @{gOMEGA - 147.4}
# Make the new phi dihedral = gPHI (nominally -57)
rotateselected {atomno = nn} {atomno = @{nn+1}} @{gPHI - 8.7}
# Make the old psi dihedral = gPSI (nominally -47)
select atomno < @{pn+2} && atomno != @{pn+3}
rotateselected {atomno=@{pn+1}} {atomno=@{pn+2}} @{gPSI + 33.4 + pb}
}
# Step new and previous N
pn = nn
nn = gN
# Make the peptide bond
connect
}
# Clean up
connect ([UNK].CA) ([UNK].Xx and within(group, _1))
select all
fix none
print format("%d atoms generated", gN)
}
echo Generating Alpha Helix
# Get the sequence from the user
gSeq = prompt("Enter AA sequence (1 char coded)", "")%9999%0
if (gSeq.count > 0) {
if (gSeq[2] == ':') {
gCHAIN = gSeq[1]
gSeq[1] = ' '
gSeq[2] = ' '
gSeq = gSeq%0
}
print format ("Sequence=%s phi=%d psi=%d", gSeq, gPHI, gPSI)
print format ("chain=%s peptide angle=%d pro bump=%d", gCHAIN, gPEPTIDE_ANGLE, gPRO_BUMP)
genAlpha(gSeq)
}
Starting on v. 11.3.15, Jmol (both application and applet) allows a customized pop-up menu instead of the standard default menu. Their definition and use are explained in Custom Menus.
Examples:
Users, if you design a new menu, please, share it here.
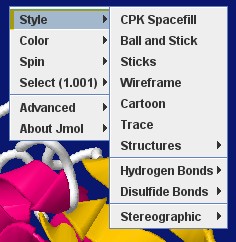
SimpleBio
This is devised to ease the handling of the menu by inexperienced users with only basic tasks, in tutorial pages of simple biomolecules.
It includes just these entries:
- Style - with simplified contents.
- Color - compacted to have less submenu nesting.
- Spin - just with on/off, no axes or speed options.
- Select - a bit simplified.
- Advanced (*) - a new entry including:
- Choose menu (*) - that allows to revert to the standard full menu.
- Language
- Console
- About
(*) These are new entries, and so will not be localized; you may wish to edit them for your intended language.
You can get this custom menu here.
SimpleChem
This is devised to ease the handling of the menu by inexperienced users with only basic tasks, in tutorial pages of organic/inorganic molecules (no crystallography, symmetry, vibration or animation, no pdb-related options).
It includes just these entries:
- Style - with simplified contents.
- Color - simplified.
- Spin - just with on/off, no axes or speed options.
- Select - a bit simplified.
- Measurements
- Advanced (*) - a new entry including:
- Choose menu (*) - that allows to revert to the standard full menu.
- Language
- Console
- About
(*) These are new entries, and so will not be localized; you may wish to edit them for your intended language.
You can get this custom menu here.
Contributors
AngelHerraez, Ceroni, Remig, Tstout, EricMartz, NicolasVervelle, Pimpim, Gutow